Yan-Yeung Luk

Yan-Yeung Luk
Associate Professor
CONTACT
Chemistry
3-048 Center for Science and Technology
Email: yluk@syr.edu
Office: 315.443.7440
PROGRAM AFFILIATIONS
Biochemistry
Biotechnology
Degrees
- B.S., 1994, Polytechnic University
- Ph.D., 2001, University of Chicago
- Postdoctoral Fellow, University of Wisconsin-Madison
Social/Academic Links
Courses Taught
- CHE 117: General Chemistry Laboratory
- CHE 275: Organic Chemistry Lecture I
- CHE 276: Organic Chemistry I Laboratory
- CHE 326: Organic Chemistry II Laboratory
- CHE 400/600: Chemistry at the Interfaces – Biomaterials
- CHE 427/627: Intermediate Organic Chemistry
- CHE 685: Organic Mechanisms
Bioorganic chemistry; surfaces; nanomaterials; chemical biology
Organic synthesis of bioactive molecules for controlling microbial lifestyles.
We conduct extensive synthesis of small organic molecules with no cytotoxicity towards mammalian cells, but can alter the bioactivities of microbes, including killing them without invoking drug tolerance, persistence, and resistance. We often design structures based on structural activity relationship (SAR) studies, and on discovering their receptor protein. Our current efforts involve carbohydrate chemistry and constructing hydrocarbons with precise branched substituents to achieve targeted control over bacterial activities.
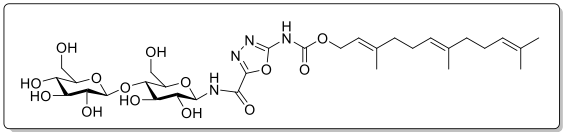
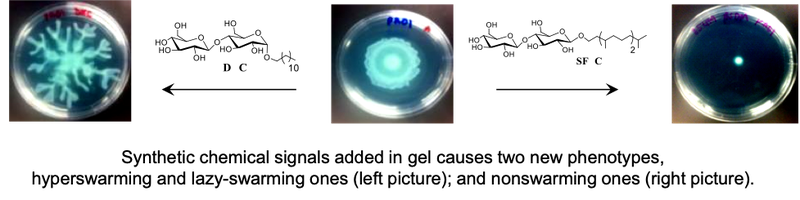
Chemical biology of bacterial signaling and development of drug tolerance, persistence, and resistance.
Bacteria use protein appendages on their surfaces to sense cues from environment and transduce such signals to trigger or to suppress protein expression and/ or small messenger molecule synthesis inside the bacteria, resulting in different bioactivities and phenotypes. Certain, actually many, small organic molecules, if one is lucky enough to discover them, function as drugs for bacteria that will alter the bacterial signaling. Some of these molecules are important because some of the signaling pathways govern the development of drug tolerance, and drug persistence, that can be happen within a matter of minutes to hours. We have found molecules that cause the bacteria to turn into two new populations with phenotypes different from each other, and from the old and abandoned phenotypes. We have discovered molecules that suppress drug tolerance and persistence, and block the transfer of resistance genes.
Engineering of Label-free bioassays for discovering new ligand-receptor interactions.
One of the wonders of life is the tight and selective binding between two biomolecules (small molecule and protein, or protein-protein binding). These binding entails the working a drug, the infection of a virus, as well as the signaling events for a bacterium or living beings in general. The validation of such binding events often requires chemical modification of one of the molecules with some sort of labels, or to immobilize one of them on a surface, in order to detect their interactions. These modifications inherently alter the binding event and can also cause false negative results. Here, we have found a way to use bacteria’s ability to senses its environment and are developing a bacterial motility enabled binding (BMED) assay that can detect a wide range of certain small molecule-protein binding interactions without any chemical labels. When the small molecules are hydrophobic, there are often nonspecific weak binding to proteins in general. This bacteria motility enabled binding assay can overcome the nonspecific binding and report only selective interaction between the ligand and the protein receptor.
- Stereochemical Society of Greater New York, Travel Award, 2007
- NSF CAREER, 11-1-2009 to 10-31-2014
- Scientific Advisory Board, Orthobond Inc. 2011-
- Non-Amphiphilic Assembly in Water: Polymorphic Nature, Thread Structure and Thermodynamic Incompatibility Lei Wu, Jyotsana Lal, Karen A. Simon, Erik A. Burton and Yan-Yeung Luk* J. Am. Chem. Soc., 2009, 131, 7430-7443.
- Water-Driven Chemoselective Reaction of Squarate Derivatives with Amino Acids and Peptides Preeti Sejwal, Yongbin Han, Akshay Shah and Yan-Yeung Luk*, Org. Lett., 2007, 9, 4897-4900. » full article
- Enhancing Cell Adhesion and Confinement by Gradient Nanotopography Karen A. Simon, Erik A. Burton, Yongbin Han, Jun Li, Anny Huang and Yan-Yeung Luk*, J. Am. Chem. Soc., 2007, 129, 4892-4893.» full article
- A Biocompatible Surfactant with Folded Hydrophilic Head Group: Enhancing the Stability of Self-Inclusion Complexes of Ferrocenyl in a β-Cyclodextrin Unit by Bond Rigidity Yongbin Han, Kejun Cheng, Karen A. Simon, Yanmei Lan, Preeti Sejwal, Yan-Yeung Luk*, J. Am. Chem. Soc., 2006, 128, 13913-20. » full article
- Surface-Driven Switching of Liquid Crystals using Redox-Active Groups on Electrodes Yan-Yeung Luk, Nicholas L. Abbott,* Science, 2003, 301, 623-6.
(Aug. 17, 2016)
Biologist Anthony Garza, Chemist Yan-Yeung Luk will 'clone' organic compounds from unculturable bacteria
(Nov. 10, 2015)
Yan-Yeung Luk's Research Group develops therapeutic agents from synthetic molecules, which mimic, dominate ones made from bacteria
(Sept. 4, 2015)
Professors Yan-Yeung Luk, John Chisholm redefine possibilities of organic chemistry
